Abstract
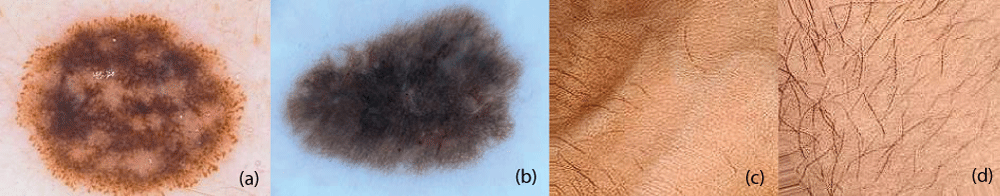
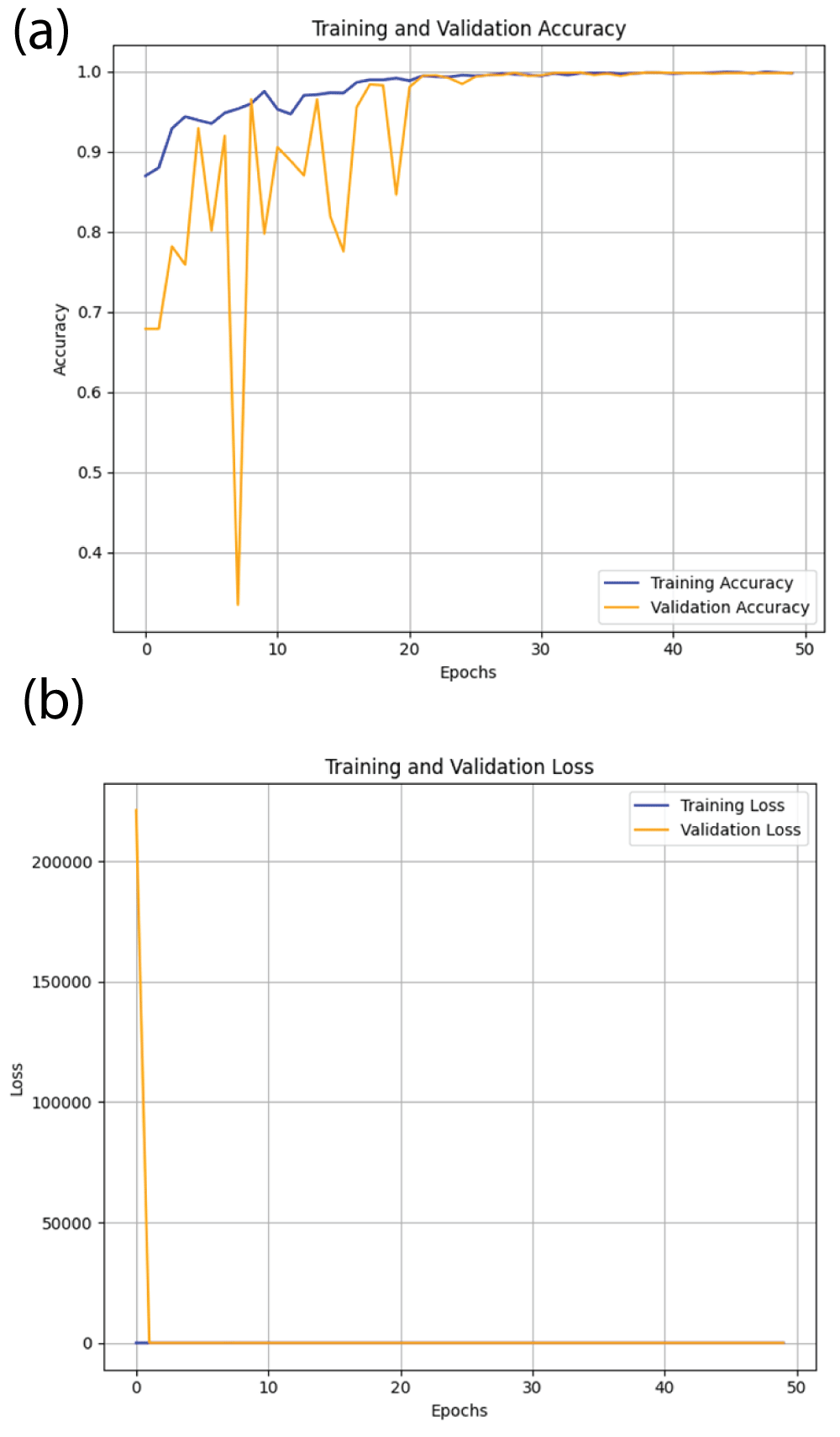
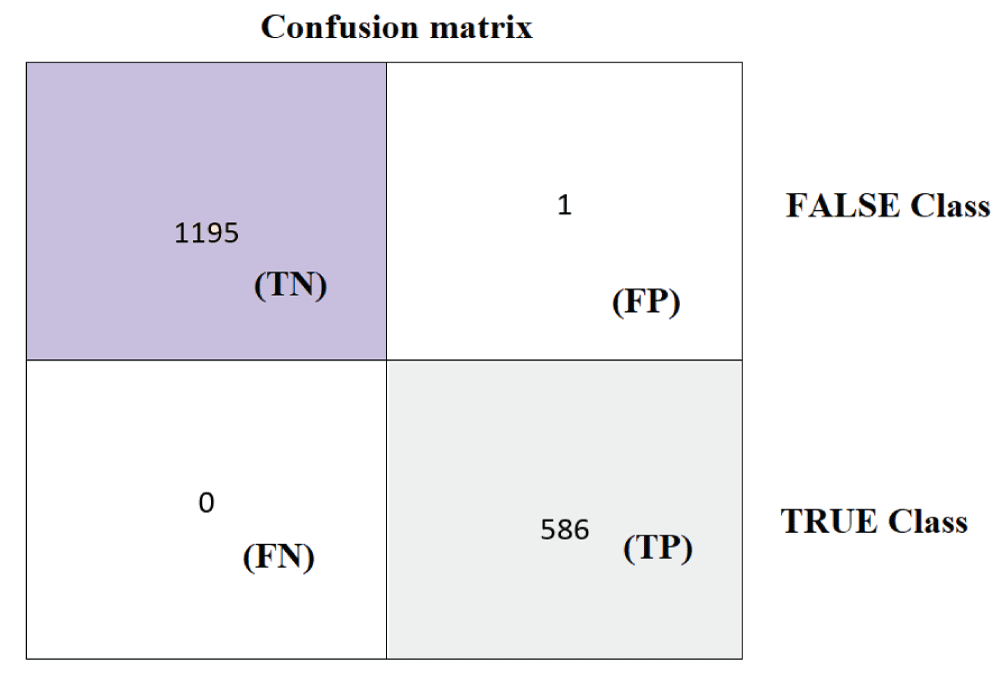
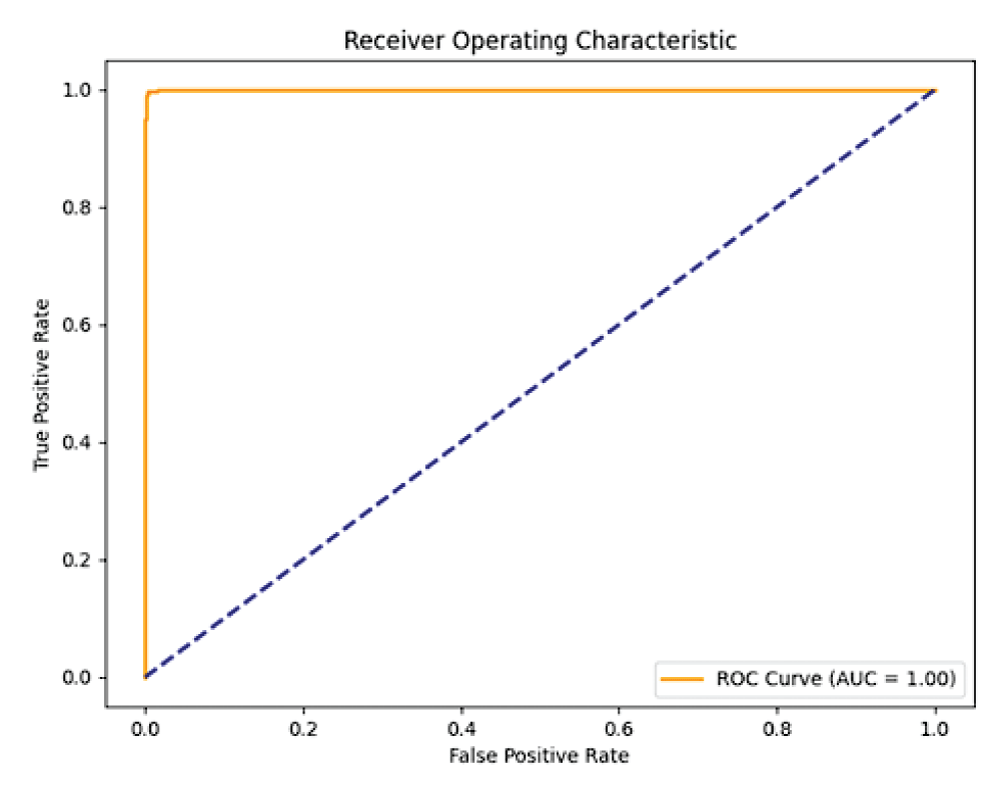
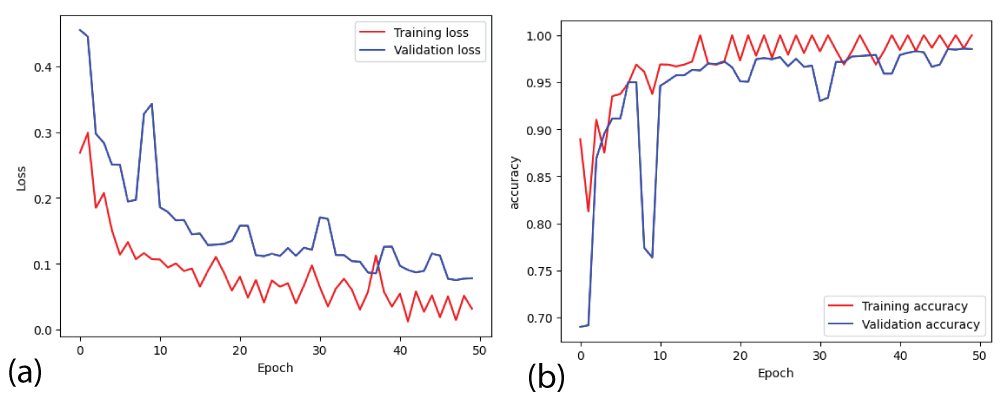
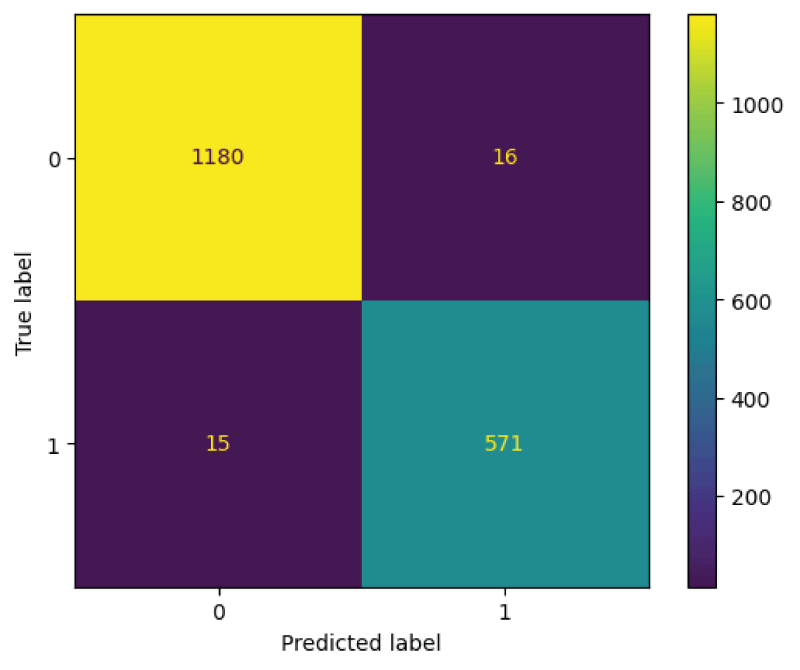
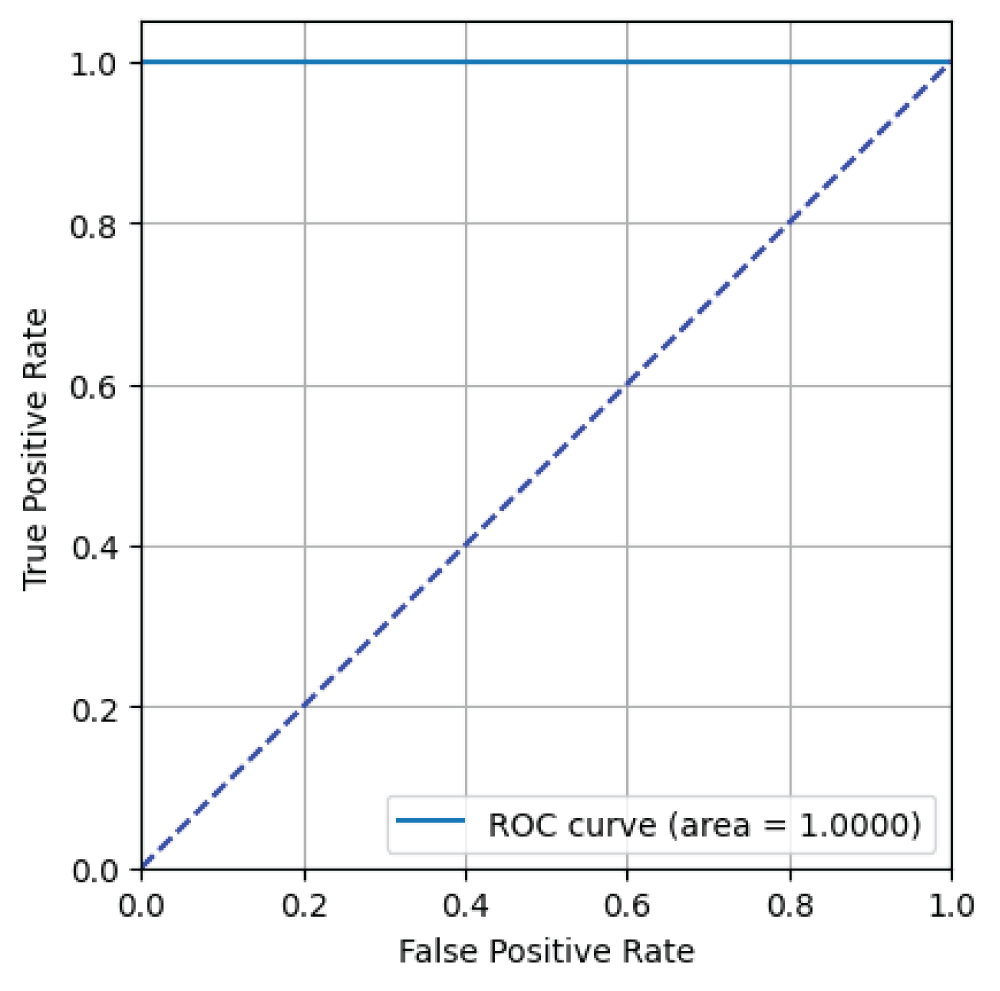
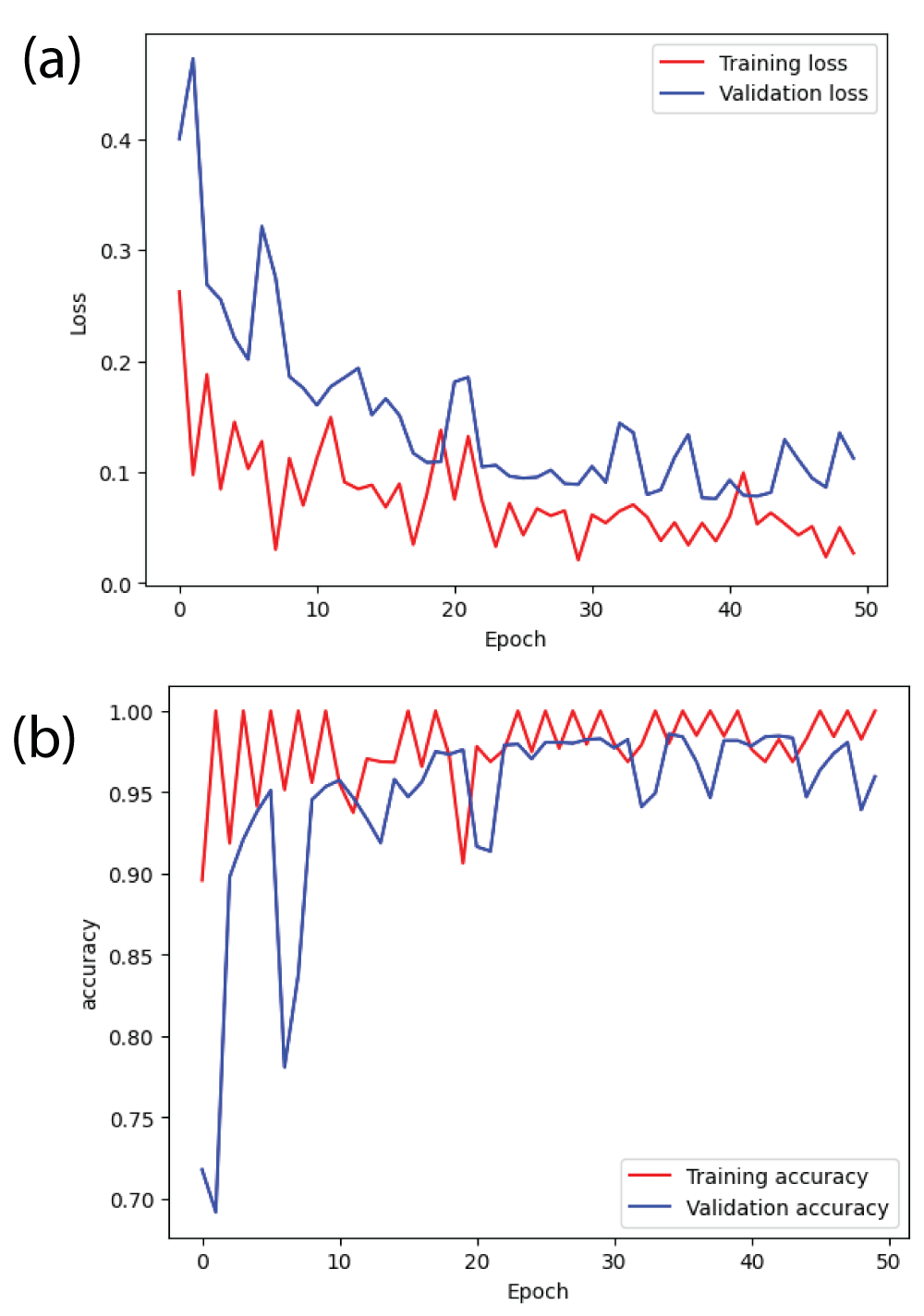
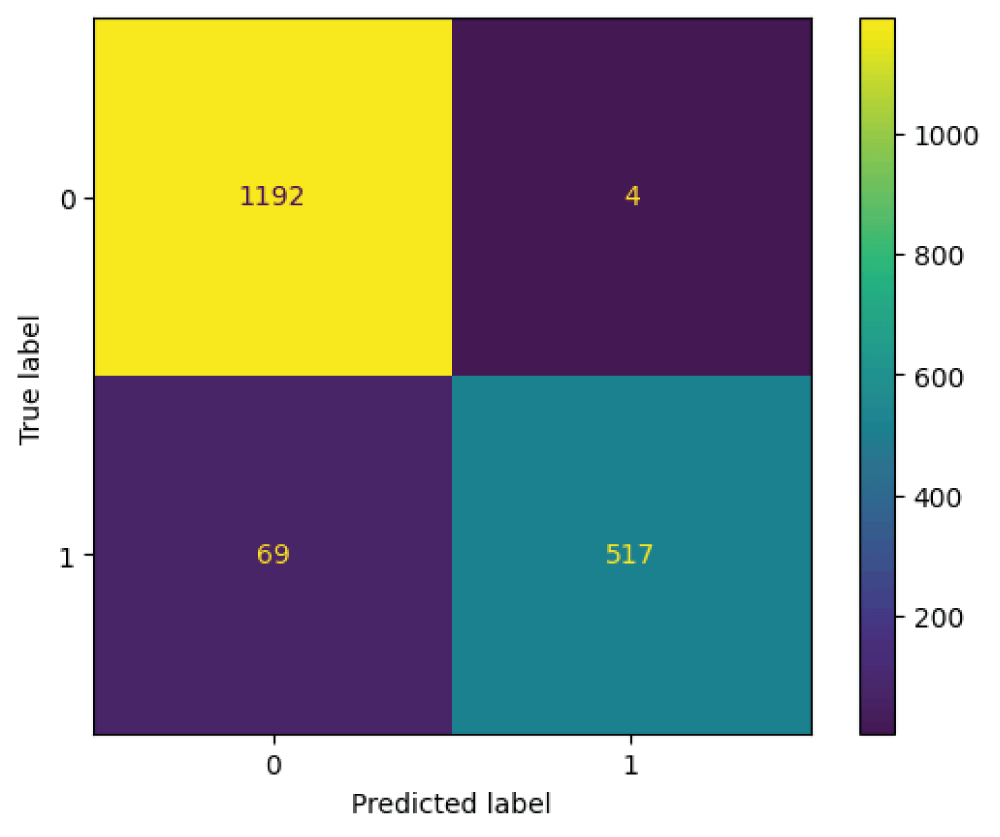
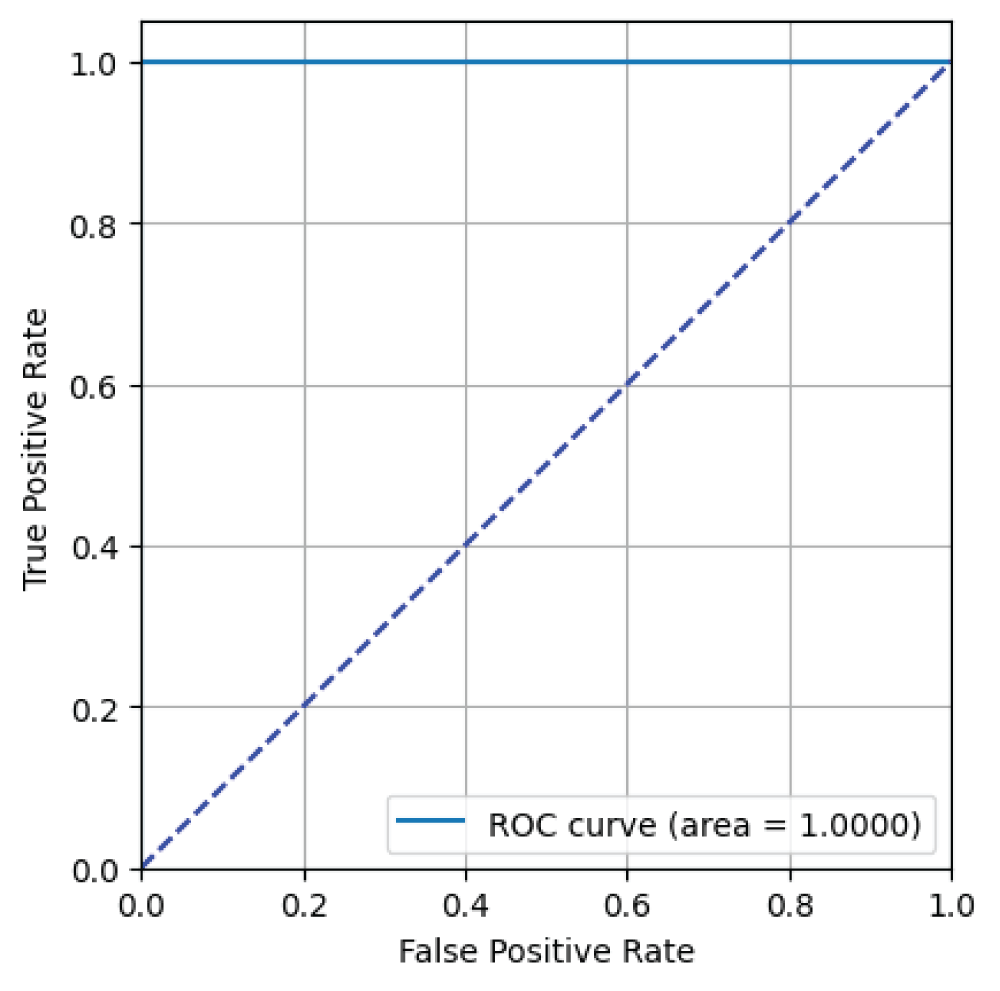
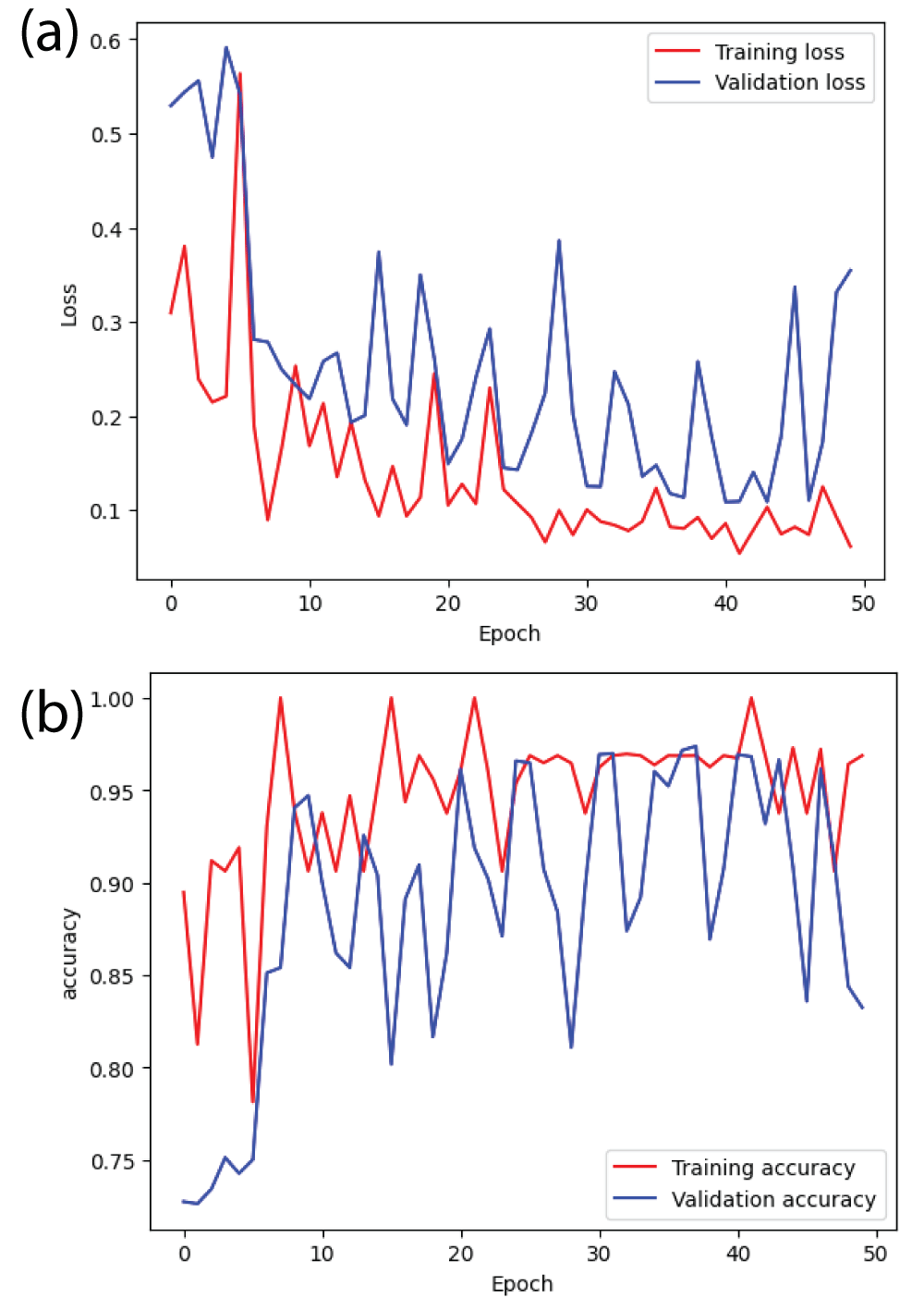
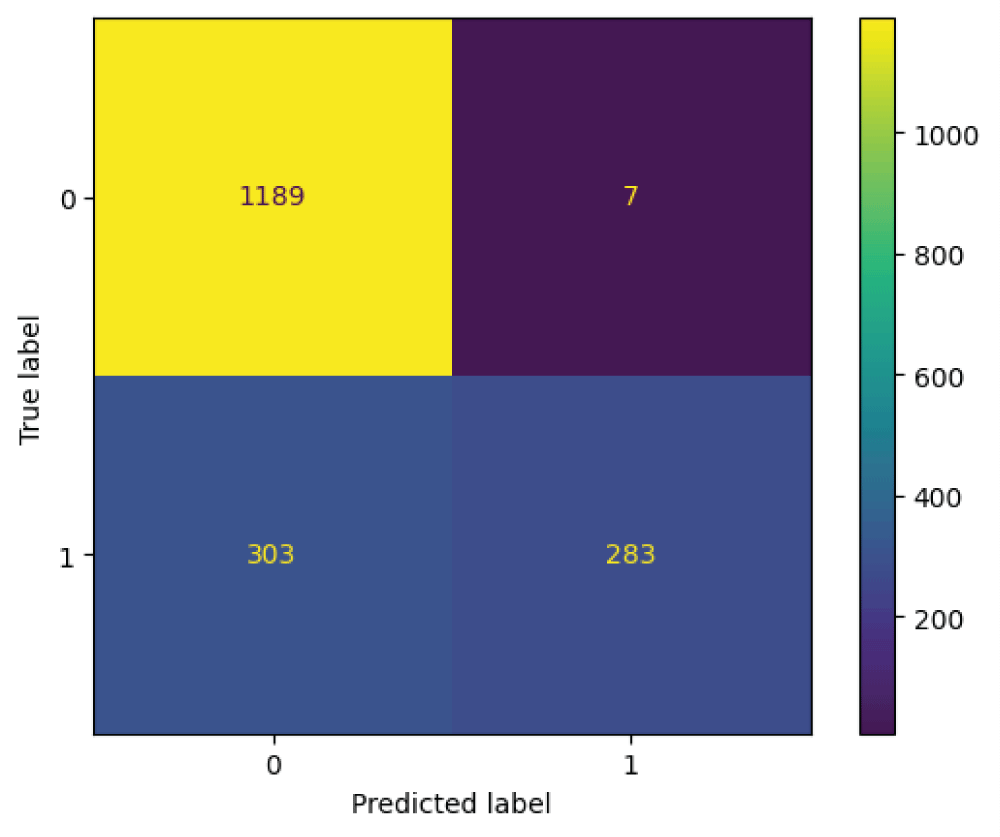
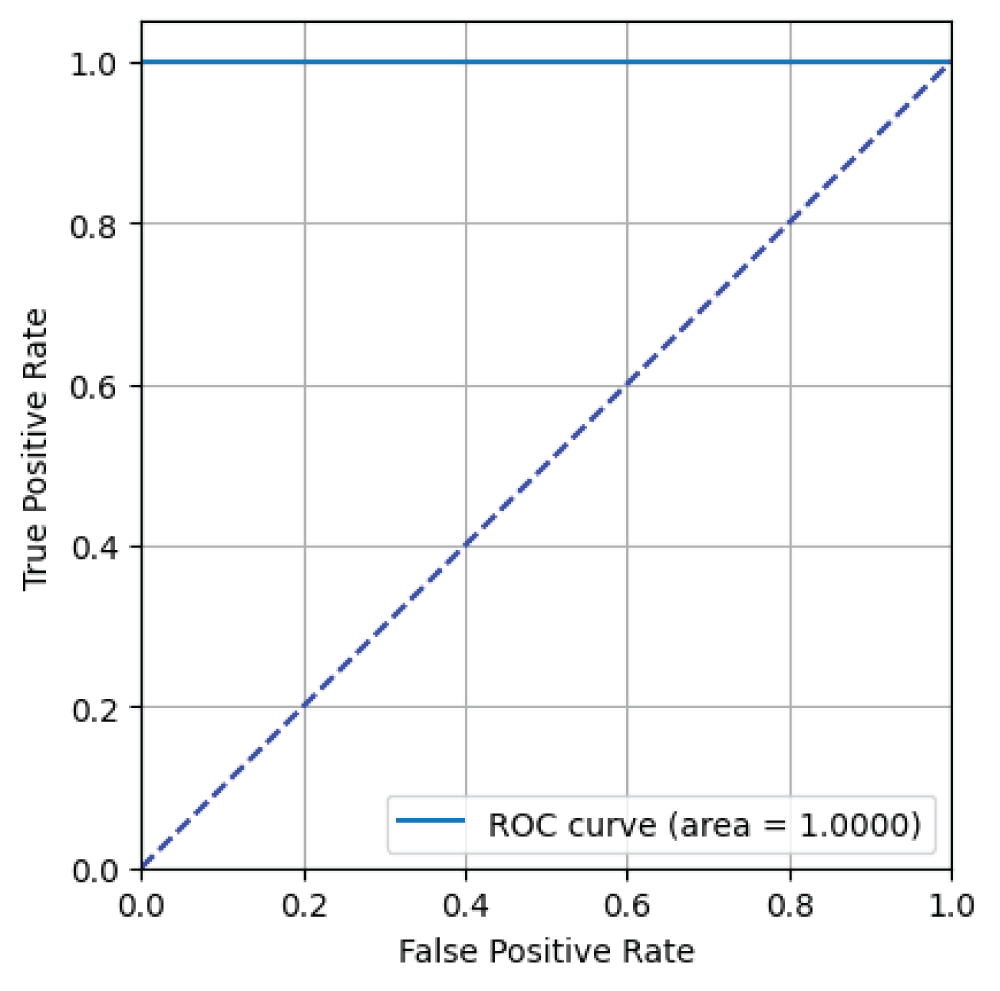
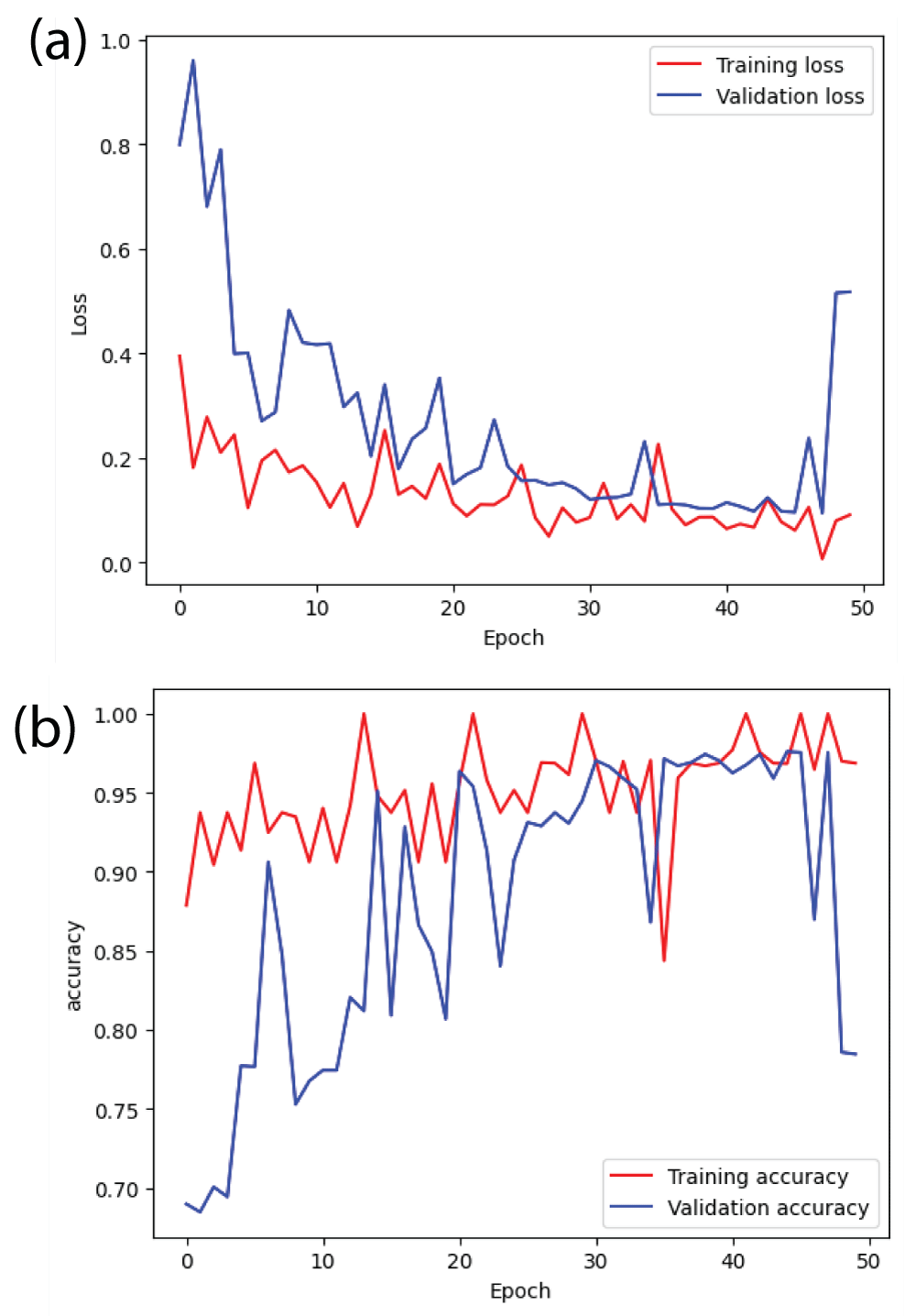
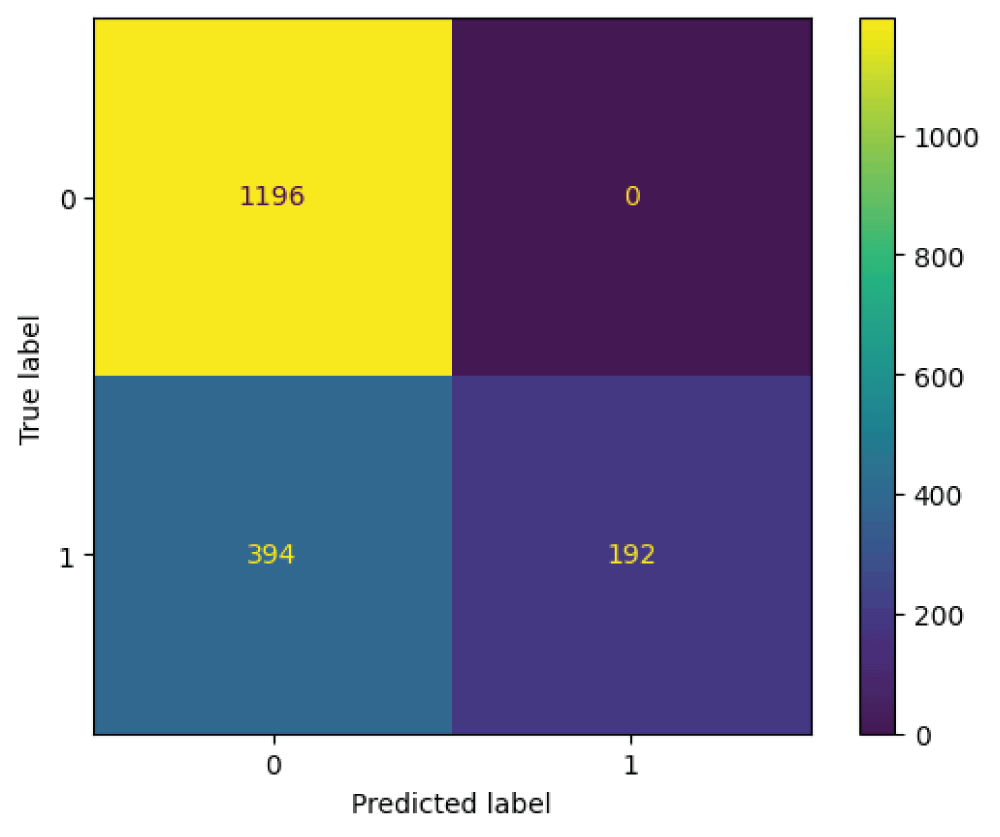
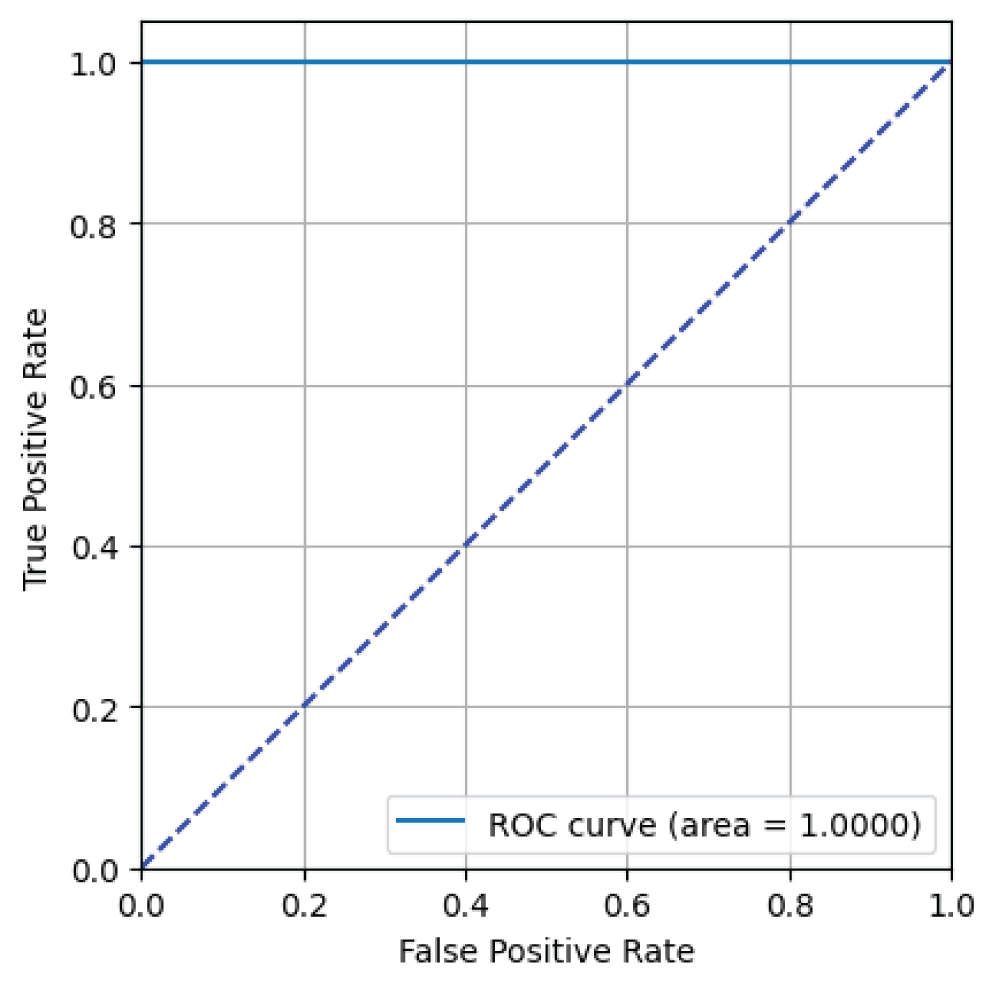
In this paper, the binary classification of skin images has been performed using deep learning technique. i.e the skin disease recognition has been performed using deep learning technique. Here, the binary classification of skin images namely melanocytic nevi and normal skin images has been classified using resnet50 deep learning network. Normal skin images have been considered in TRUE class. Melanocytic nevi skin images have been considered in FALSE class. Traditional method such as biopsy involves lot of computational procedures and consumes a lot of time which is tedious process. Therefore, deep learning-based skin disease recognition has been proposed here. The dataset for melanocytic nevi and normal skin images have been prepared for 9792 images. This dataset is passed through all five deep residual network models to obtain the results. The results such as error/accuracy curves, error matrix, false-positive-rate (FPR) vs. true-positive-rate (TPR) curve are shown for the confirmation of the work. The results obtained from the ResNet50 model were compared with other deep residual network models i.e ResNet18, ResNet34, ResNet101, and ResNet152 models.